Finding Bacteria in Blood
A Hardware-based Neuromorphic Solution for Real-World E-Nose Applications
Executive Summary:
Advancements in neuromorphic technology, which mimics the human brain has opened exciting possibilities in creating machines that can smell, much like our noses. This paper introduces a bio-inspired, low-power method that recognizes different patterns of scents, functioning as the “brain” of an electronic nose. This highly efficient and compact solution can be seamlessly incorporated into electronic nose systems targeted for high-precision, real-world applications. In practical tests, it has proven very effective, especially in swiftly identifying various bacteria in blood samples using electronic nose data. The proposed method achieves 181 inferences per second with a classification accuracy of 97.42% and dynamic efficiency, measured as energy consumed per inference, of 135 µJ/inference. This combination of accuracy, efficiency, and speed makes it an exceptional tool for health and safety applications that can be used in a cost-effective, portable form factor in remote regions, which may not have access to labs or even connectivity to the cloud.
Advancing Artificial Olfaction: Bridging Biology and Technology
Electronic noses (e-noses) serve a vital role in applications such as quality assurance in food production and non-invasive medical diagnostics. They are sophisticated devices designed to detect odours. They operate by mimicking the biological process of smell, where specific neurons in the nose identify volatile organic compounds and send encoded signals to the brain for odour recognition. Electronic noses are equipped with an array of chemical sensors that produce electrical signals when they encounter airborne molecules, known as Volatile Organic Compounds (VOCs). The unique pattern of these signals is akin to a fingerprint, which can be analysed and identified using various techniques to determine the specific odour present.
Despite the technological prowess demonstrated in laboratory settings, the use of electronic noses in everyday environments faces challenges. Limitations include the need for greater portability, improved power efficiency, enhanced performance, and quicker, more reliable odour classification. Advances in machine learning have increased the accuracy of electronic noses; however, these improvements often come at the cost of increased computational demand, affecting the device’s portability and energy usage. Current research in neuromorphic computing, inspired by the brain’s efficient processing, promises to overcome these limitations by emulating the accuracy and low-energy profile of the natural olfactory system. While previous studies have achieved intricate biological mimicry, such complexity is not practical for widespread application.
Our research presents a bio-inspired approach that harnesses the efficiency of neuromorphic hardware. We have developed a neuromorphic system that processes electronic nose data using a combination of an event-based data encoder and a spiking neural network classifier optimized for implementation on Akida neuromorphic hardware. This system is built to leverage the inherent strengths of neuromorphic computing, aiming to provide a robust inference engine capable of high-accuracy odour classification with low power consumption and minimal delay, ready for real-world deployment.
Dataset Overview:
In this research, we utilized the ‘bacteria in blood’ dataset, which was previously collected by the Mednose project team at Örebro University. Their project was centred on the early detection of different bacterial species in blood, by identifying the unique chemical signatures they emit during initial growth phases. The collection was carried out using the NST 3220 Emission Analyzer, a sophisticated electronic nose device developed by Applied Sensors, equipped with a combined array of 22 metal-oxide semiconductor (MOS) and MOSFET sensors. The project’s objective was to distinguish between ten types of bacteria in blood samples, and for this purpose, a comprehensive dataset of 1200 samples was created, with 120 samples representing each bacterial species. For an in-depth understanding of the electronic nose experiments and the sampling protocols, a detailed description is available in the work by Trincavelli et al. titled “Direct identification of bacteria in blood culture samples using an electronic nose,” published in the IEEE Transactions on Biomedical Engineering
Streamlining Data: From Raw Signals to Event-Driven Patterns
Pre-processing plays a pivotal role in transforming raw complex sensor data into a more manageable form for pattern recognition and analysis. We start by removing any constant background noise from the readings—a step known as baseline cancellation. Then we adjust the scale of the sensor responses to ensure consistency across all samples.
The heart of our approach is a specialized model designed to interpret these streamlined signals. Our model is fine-tuned for data that’s been transformed into a pattern of digital events, similar to the way our brain receives and processes sensory input. To convert the sensor’s continuous data into this digital format, we explored two methods. The first method, based on our previously published encoding algorithm known as Address Event Representation for Olfaction (AERO), arranges the sensor information into a structured grid, marking the presence of a signal at each moment in time. This is akin to creating a digital snapshot of the scent at every point, which helps us capture the essential characteristics of each odour.
The second method, the Step Forward (SF) algorithm, a bio-inspired data encoding method, highlighting changes in the odour data only when they pass a certain threshold. This is inspired by how our senses work, paying attention to changes rather than static information, making it efficient and effective at capturing rapid variations in odour signals.
We also considered that not all data might be necessary for accurate identification. Perhaps, after the initial burst of information when the scent is first detected, the remaining data might not add much value. So, we experimented with using different amounts of data to see if we could still identify the odours without the full 5-minute recording, focusing only on the critical moments that define each scent.
Optimising Akida Spiking Neural Networks for Effective Odour Detection
The core of our classification system is a spiking neural network (SNN), which mimics the way neurons in the brain process information and learn. It consists of two main layers: the input layer, where data comes in, and the processing layer, where the learning happens. The neurons within the processing layer are finely tuned to learn and recognize patterns in the incoming data and respond by firing when a familiar pattern is identified, thereby signalling the detection of a specific odour.
The learning mechanism of our SNN is rooted in a biological process known as Spike Time Dependent Plasticity (STDP). This natural learning mode allows the network to rapidly strengthen the synaptic connections that correspond to frequently encountered activation patterns, a fundamental aspect of how the brain learns from repetition and reinforcement. In the classification phase, the system employs a competitive process where the most activated neuron—indicating the recognition of a learned pattern—determines the odour’s classification. This is achieved through the winner-take-all approach, where the ‘winning’ neuron’s response guides the output classification.
To achieve the most accurate and efficient performance, we conducted a comprehensive optimisation of the network’s hyper-parameters. This involved adjusting the number of neurons associated with each class of odour, the number of active connections each neuron should have, and the intensity of learning competition among neurons. The configuration resulted in a robust model that could accurately classify odours based on the patterns it had learned.
The SNN model was rigorously tested and validated using MetaTF, a Python-based simulation environment for AKD1000 reference SoC, before its deployment on the Akida reference hardware. This validation ensured that our approach was sound and could be effectively transferred to the NSoC, allowing for a seamless transition from theory to application. The entire workflow, from the initial data pre-processing to the final odour classification, is designed to be fully compatible and integrated within the NSoC, showcasing the system’s readiness for deployment in electronic nose devices.
Performance Evaluation of Akida SNN Classifier
The objective of this study was to evaluate whether the spiking neural network (SNN) could accurately classify odours without needing to process the entire 5-minute sample, thus potentially reducing the time for analysis. We used a windowed approach, where the electronic nose data and then converted into an event-based format by the data-to-event encoder for the SNN to process.
Our performance assessment of the SNN classifier utilised a robust 3-fold cross-validation method, ensuring a reliable statistical evaluation. The results, summarized in the classification table, showed high mean classification accuracies for both encoding algorithms (AERO and Step Forward), particularly with the first 200 data points. This suggests that the essential features for odour differentiation are present early in the sampling process.
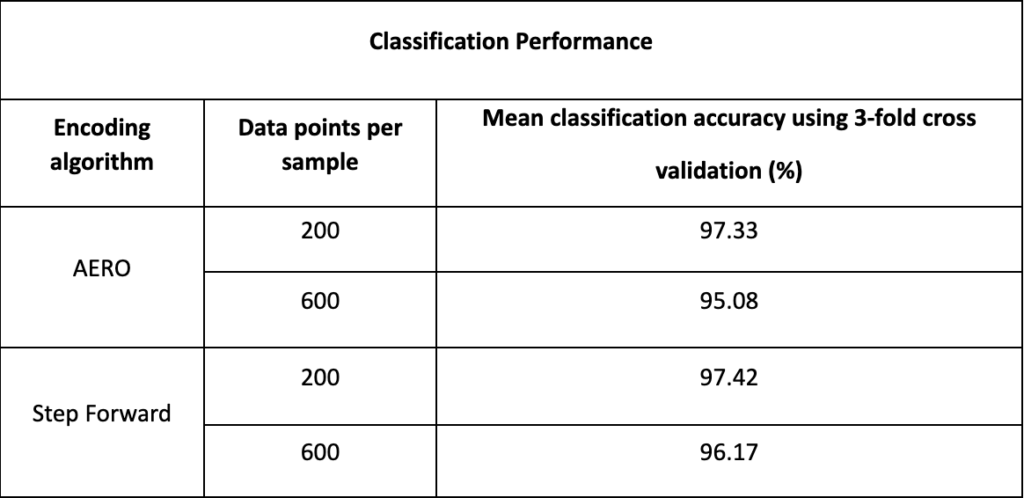
The Step Forward encoding technique, while similar in classification accuracy to AERO, was more data-efficient, condensing the information into a smaller input size for the SNN. However, this efficiency required additional pre-processing, which could slightly increase the time to reach a classification result.
For real-world application, the classifiers were tested on the Akida Neuromorphic System-on-Chip (NSoC). The SNN classifiers performed on par with the MetaTF chip emulator used during development. The AERO-based classifier model demonstrated a dynamic power consumption of only 24.5 mW with a high throughput of 181 inferences per second, translating to an energy efficiency of 135 µJ per inference. On the other hand, the Step Forward encoded model consumed slightly more power, averaging 25 mW, with a lower throughput of 31.5 inferences per second, resulting in a dynamic efficiency of 822 µJ per inference.
These differences in performance and efficiency are attributed to the varying neural resources each model utilized. Nonetheless, the findings affirm the potential of SNN classifiers to function as low-power, real-time pattern recognition engines for electronic nose systems, providing a balance of speed, accuracy, and energy efficiency.
Revolutionizing Diagnostic Tools: Akida’s NSoC-Powered E-Nose

Figure 1: Shows the Mednose apparatus with Applied Sensor NST320 and attached computer.
The system used to develop this Mednose system is fixed, large, power-hungry and needs a great deal of compute. While there are portable systems available for the sensing, they are not capable of doing the diagnostics on device today.
The quest for a real-time, energy-efficient system for odour recognition has long stood as a critical challenge in the development of electronic nose technologies. This paper has showcased a significant leap forward in this domain: the application of an event-based neuromorphic approach for processing and classifying electronic nose data, marking a substantial innovation in pattern recognition engines for e-nose systems.
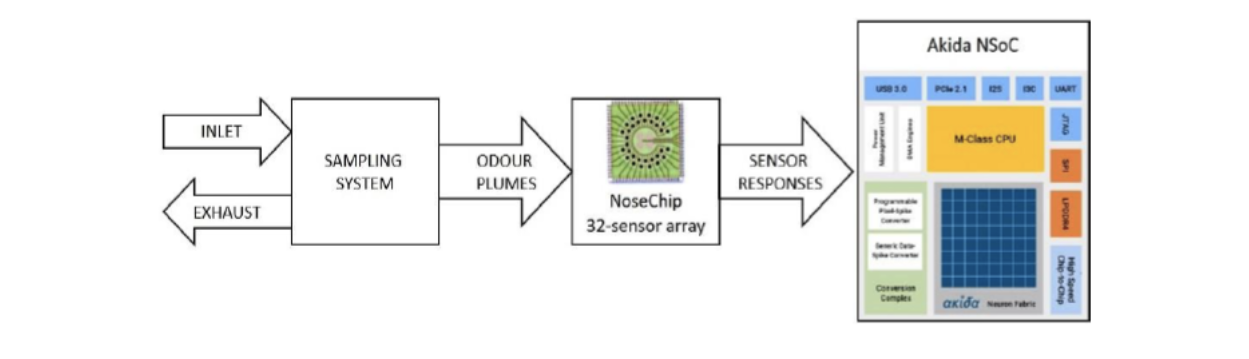
Fig 2: Sample diagnostic platform using NoseChip 32-array sensor and AKD1000 SoC which can enable single board, extremely compact, energy-efficient, and accurate solution.
In summary, the proposed solution integrates two key components: an innovative data-to-event encoder using AERO and Step Forward (SF) techniques for translating sensor data into a neural-compatible format and an Akida SNN classifier that employs biologically inspired algorithms to discern and categorize patterns in the data.
The performance of this system is notable—when tested on the MedNose dataset, it demonstrated an impressive ability to distinguish among ten different bacteria types, achieving a peak classification accuracy of 97.42%. This not only surpasses previous models, which needed multiple samples to reach slightly lower accuracy levels, but also does so with remarkable efficiency. Critically, the SNN model was rigorously tested on the Akida NSoC, where it exhibited exceptionally low power usage, consuming only 24.5 mW, while maintaining a high throughput of 181 inferences per second. This performance marks a significant advancement, indicating that such a system is not only possible but now can be provided with a very cost-effective and portable device that enables rapid disease diagnosis with the critical level of precision.
These results highlight the processing capabilities of the Akida SNN for complex time-varying signals and demonstrate potential impact of our company’s technology in real-world applications. When used in conjunction with such sensing systems, the implications for healthcare are particularly profound, where these advancements could drastically reduce the time and resources required to assist in disease detection and identification, ultimately leading to faster, more accurate medical responses and better patient outcomes.
Click here to read the White paper for more details.




